W.B.Langdon . 6 October 2015 2014 papers , full list
W. B. Langdon, Technical Report RN/14/13
ABSTRACT
Discussion of the state of the art in and future research on Software Testing during NII Shonan Meeting Computational Intelligence for Software Engineering, Seminar 053, 20-23 October 2014. The discussions between software engineers and experts in artificial intelligence were mainly lead by Andreas Zeller and Jens Krinke






W. B. Langdon,
BioData Mining,
2014
7(3).
Draft
doi:10.1186/1756-0381-7-3
![]()
![]()
Background
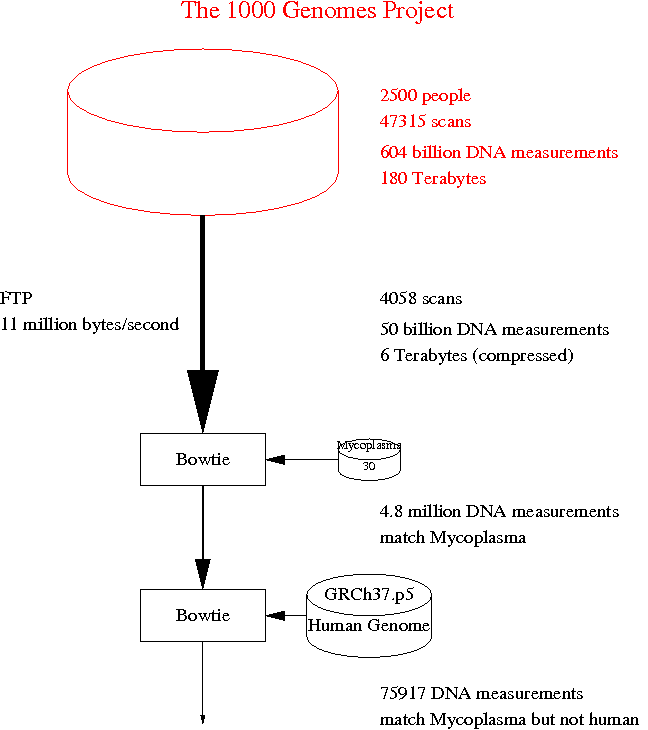
In silco Biology is increasingly important and is often based on public data. While the problem of contamination is well recognised in microbiology labs the corresponding problem of database corruption has received less attention [1 More Mouldy Data In Silico Infection of the Human Genome].
Results:
Mapping 50 billion next generation DNA sequences from The Thousand Genome Project against published genomes reveals many that match one or more Mycoplasma but are not included in the reference human genome GRCh37.p5. Many of these are of low quality but NCBI BLAST searches confirm some high quality, high entropy sequences match Mycoplasma but no human sequences.
Conclusions:
It appears at least 7% of 1000G samples are contaminated.
Keywords
Molecular Biology, Microbiology, genetics, metagenomic, Data mining, Next-generation DNA sequencing, Data cleansing, High Throughput, Solexa, 454, SOLiD.
 Endosymbiotic origin and differential loss of eukaryotic genes,
Chuan Ku, Shijulal Nelson-Sathi, Mayo Roettger, Filipa L. Sousa, Peter J. Lockhart, David Bryant, Einat Hazkani-Covo, James O. McInerney, Giddy Landan & William F. Martin,
Nature 524,
427-432
doi:10.1038/nature14963
PMID: 26287458
Endosymbiotic origin and differential loss of eukaryotic genes,
Chuan Ku, Shijulal Nelson-Sathi, Mayo Roettger, Filipa L. Sousa, Peter J. Lockhart, David Bryant, Einat Hazkani-Covo, James O. McInerney, Giddy Landan & William F. Martin,
Nature 524,
427-432
doi:10.1038/nature14963
PMID: 26287458
 Using populations of human and microbial genomes for organism detection in metagenomes,
Sasha K. Ames, Shea N. Gardner, Jose Manuel Marti, Tom R. Slezak, Maya B. Gokhale, Jonathan E. Allen,
Genome Res. 2015 July; 25(7): 1056-1067.
doi: 10.1101/gr.184879.114
PMCID: PMC4484388
Using populations of human and microbial genomes for organism detection in metagenomes,
Sasha K. Ames, Shea N. Gardner, Jose Manuel Marti, Tom R. Slezak, Maya B. Gokhale, Jonathan E. Allen,
Genome Res. 2015 July; 25(7): 1056-1067.
doi: 10.1101/gr.184879.114
PMCID: PMC4484388
![]() Large-scale contamination of microbial isolate genomes by Illumina PhiX control.
Supratim Mukherjee, Marcel Huntemann, Natalia Ivanova,
Nikos C Kyrpides
and Amrita Pati,
Standards in Genomic Sciences 2015,
10:18
PMCID: PMC4511556
Large-scale contamination of microbial isolate genomes by Illumina PhiX control.
Supratim Mukherjee, Marcel Huntemann, Natalia Ivanova,
Nikos C Kyrpides
and Amrita Pati,
Standards in Genomic Sciences 2015,
10:18
PMCID: PMC4511556
A Novel Method for Detecting Contaminated Sample Based on Illumina Sequencing Data. International Journal of Bioscience, Biochemistry and Bioinformatics, 4(2) 2014, Zheng Huang and Qibin Li and Wei Jin and Qijun Liao and Xiao Sun http://www.ijbbb.org/papers/322-E0014.pdf
 Assessing the prevalence of mycoplasma contamination in cell culture via a survey of NCBI's RNA-seq archive,
Anthony O. Olarerin-George, John B. Hogenesch
Nucleic Acids Res. 2015 March 11; 43(5):
2535-2542.
doi: 10.1093/nar/gkv136
PMCID: PMC4357728
Assessing the prevalence of mycoplasma contamination in cell culture via a survey of NCBI's RNA-seq archive,
Anthony O. Olarerin-George, John B. Hogenesch
Nucleic Acids Res. 2015 March 11; 43(5):
2535-2542.
doi: 10.1093/nar/gkv136
PMCID: PMC4357728
![]()
![]() Here, there, and everywhere: From PCRs to next-generation sequencing technologies and sequence databases, DNA contaminants creep in from the most unlikely places,
Karl Gruber,
EMBO Rep. 2015 August; 16(8):
898-901
PMID: 26150097
Here, there, and everywhere: From PCRs to next-generation sequencing technologies and sequence databases, DNA contaminants creep in from the most unlikely places,
Karl Gruber,
EMBO Rep. 2015 August; 16(8):
898-901
PMID: 26150097