W.B. Langdon and J.P. Nordin
Centrum voor Wiskunde en Informatica,
Kruislaan 413, NL-1098 SJ, Amsterdam
Chalmers University of Technology,
S-41296, Göteborg, Sweden
nordin@fy.chalmers.se
We want to find patterns in data.
Want
generalisation on unseen data.
To find the ``essence'' of the data.
We turn genetic programming (GP) on its head.
Instead of GP matching training data.
We construct a function to match training data.
Seed GP with it.
Can GP evolve the function to be more general?

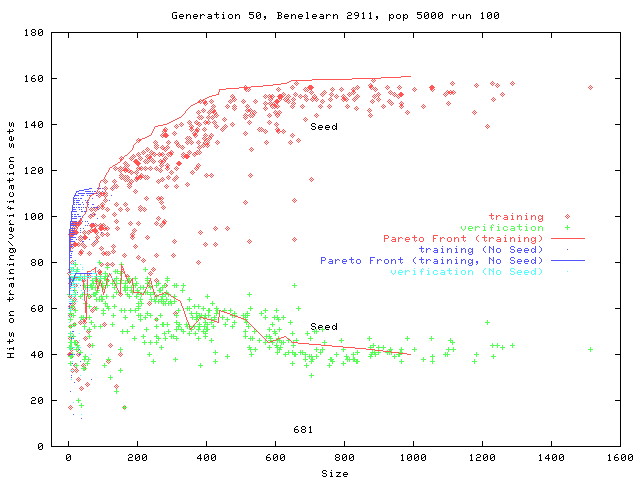
Pima Diabetes
Exactly matches training data
![]() General solution
General solutionVery simple program is as good as ML
Objective:
Find a program that predicts Diabetes.
Terminal set:
One terminal per data attribute (8).
For each attribute, create unique random constants between its minimum and
maximum value (in the training set).
Use integers where the attribute has integer only values
(total 255 primitives).
Function set:
IF IFLTE MUL ADD DIV SUB AND OR NAND NOR XOR EQ
APPROX NOT
Fitness cases:
Training 576 (194 positive).
192 (74 positive) for verification only.
Fitness:
number correct (hits).
Selection:
2 objectives, hits and size, combined in
Pareto tournament group size 7.
Non-elitist, generational. Fitness sharing (comparison set 81)
Wrapper:
Value
![]() taken as true
taken as truePop Size:
500
Max prog. size:
no limit
Initial pop:
100% seeded or
created using ``ramped half-and-half''
(no duplicate checking).
Parameters:
90% one child crossover,
2.5% function point mutation (rate 100/1024),
2.5% constant mutation (rate 100/1024),
2.5% shrink mutation,
2.5% size fair mutation (subtree size
![]() 30)
30)Termination:
Maximum number of generations G = 50
| Seed size | Highest Verification | First program |
||||
| Hits | Size | Gen | Size | Gen | % runs
|
|
| none (Pareto) | 121 | 25 | 8 | 0 | ||
| none (scalar) | 131 | 31 | 10 | 5 | 17 | 40 |
| 20 | 143 | 34 | 35 | 3 | 26 | 100 |
| 288 | 144 | 1603 | 40 | 4 | 22 | 100 |
| 576 | 142 | 6369 | 41 | 62 | 33 | 100 |
|

|
| Objective: | Find a program that predicts Breast Cancer. |
| Terminal set: | One terminal per data attribute (9). Constants -1 to 10 |
| Fitness cases: | 351 (130 positive). 348 (111 positive) verification tests. |
| Cancer. Highest verification score on Pareto front | ||||||
| Seed size | Highest Verification | First program |
||||
| Hits | Size | Gen | Size | Gen | % runs
|
|
| none (Pareto) | 333 | 23 | 28 | 6 | 14 | 100 |
| 20 | 330 | 101 | 34 | 65 | 32 | 100 |
| 128 | 327 | 46 | 39 | 42 | 38 | 80 |
| 351 | 327 | 6422 | 42 | 2622 | 47 | 80 |
Given 85 attributes relating to a customer, such as age, number of children, number of cars, income, other insurance policies they hold, predict if they want caravan insurance.
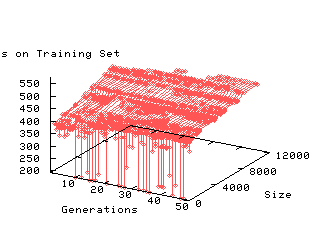
GP generalises the C4.5 seed. However over training increases with size of programs.