Predicting Cancer
W. B. Langdon,
Computer Science, University College, Gower Street, London, WC1E 6BT, UK
Data Exploration Sciences (BDS), GlaxoSmithKline Research and Development, Greenford Middlesex, UK
Computers are being used to evolve predictors based on DNA gene chips. Recently we have evolved programs that use mRNA extracted from cancerous cells to try to forecast patient outcomes.
DNA chips enable scientists to estimate how much of many proteins a cell contains. The assumption is by indirectly measuring mRNA concentration levels, DNA chips tell us about the proteins the mRNA manufactures. The excitement comes because an individual chip can indirectly measure thousands of different mRNAs and so tell us about thousands of proteins. In the near future, DNA chips will be able to tell us about every human protein simultaneously.
Of course things are not quite so easy. Firstly the chips are analogue devices, they are inevitably subject to noise. Worse they measure DNA not the proteins themselves. There are several levels of purification, transformation and amplification between obtaining the sample and taking measurements. All these are subject to noise and liable to contamination but much worse the signal strength may be different for each protein. Secondly suddenly Biologist have been swamped with an embarrassingly large volume of data. They have turned to computers to process it. Indeed Biologists and Computer Scientists have together formed a new field of Bioinformatics.
The hope is that DNA chips will lead to better understanding and hence treatment of diseases and their cures. Present research often takes DNA chip data from healthy and sick people and looks for differences between them. Particular interest is between gene expression (i.e. production of protein) in cancerous and non-cancerous cells, or between children who responded to treatment those that did not. We ask the computer to learn from the data and to find a pattern which reliably allows it to tell a healthy sample from a non-healthy sample. The pattern is of immediate use to medical research and could be used by doctors as a diagnostic aid. The learning problem is hard where the pattern is not obvious, e.g. because multiple genes are involved and/or the data are very noisy.
Machine learning has had many successes with noisy problems by learning from examples. Particular problems, are there are thousands of genes to consider and sometimes only a few training examples to learn from. Looking at every possible gene combination (even before deciding how to combine them) would mean considering a large number, say 27129, possibilities. This is not feasible.
We use computer based evolutionary search (GP) both to decide which genes to use and simultaneously to decide how they will be combined. Evolutionary search uses a population of trial predictors. Each generation the more accurate predictors are selected to have children in the next generation (c.f. survival of the fittest in Darwinian natural evolution). Each new predictor is created by copying parts of its two parents. Evolutionary search has been very successful at solving difficult combinatorial problems.
The second part of making a prediction is deciding how to combine
the chosen DNA chip data. In GP data are combined in arbitrary
mathematical functions. We leave it to evolution to decide which data
to use and how to combine them. For convience the function are stored
inside the computer as program trees (see figures).
New functions,
which will become the children in the next generation, are created by
copying branches from two parent trees and joining them together into
a new tree.
Creating new program trees
|
The programs are stored as tree structures (shown looking down on the
tree from above). DNA chip data values enters via the leafs
(gnnn). They are combined by arithmetic operations (e.g. ADD) and work
their way to the centre. A positive value means the program predicts a
positive outcome.
|
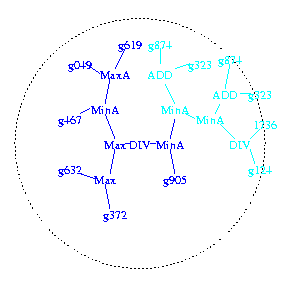
Figure 1a. First high fitness program selected to be a parent.
Code past to child in dark blue. Circle indicates 6 levels from root of tree (divide at centre of circle).
|

Figure 1b. Second high fitness program selected to be a parent. A subtree (in red) is chosen at random and also passed to child.
|
|
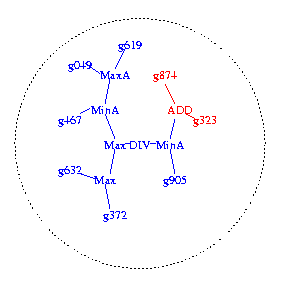
Figure 1c. New program formed by joining code fragments from both
parents.
|
UCL Science, 17, page 2, September 2003


