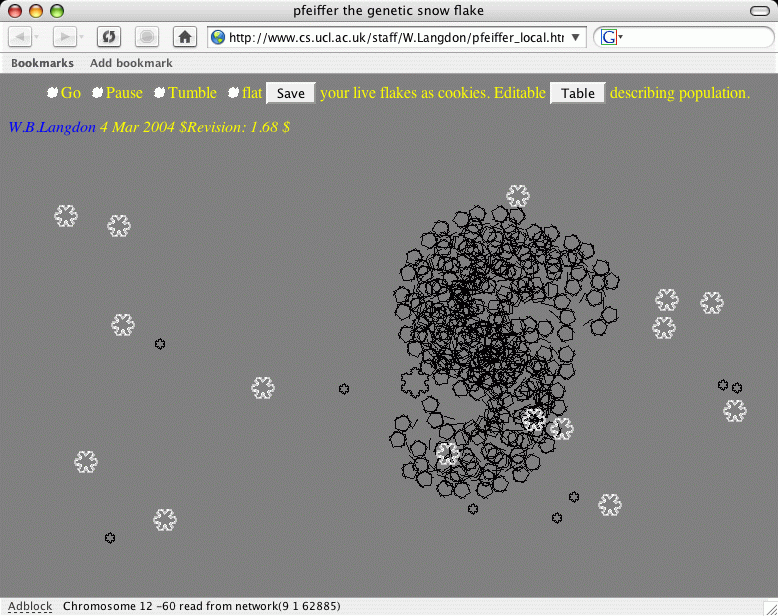
Example of main screen (pfeiffer_local.html)

The example screen shot shows a mixture of sterile initial flakes (white) and genetically determined xbm images (black). The browser started with 25 sterile flakes. It is in the process of replacing these by down loading 25 flakes from the global population of flakes previously saved by other users. It has just loaded the thirteenth (number 12) chromosome from the network and has just rendered the tenth (number 9) as xbm image. This leaves 15 white flakes to go. It can take several minutes to down load them all. (The number of flakes can be reduced, or indeed increased, by using pfeiffer_params.html before the simulation is started.) Note beauty means different things to different people. The large seahorse shape was saved by a user on 9 February 2004 at 12:57:54. The flying menus can be used to delete ugly flakes before they get a chance to breed and produce children.
By default the "Age before breeding" parameter is set sufficiently high that all flakes will be loaded before any can start breeding. This can be reduced (but should always be bigger than, say, 50).
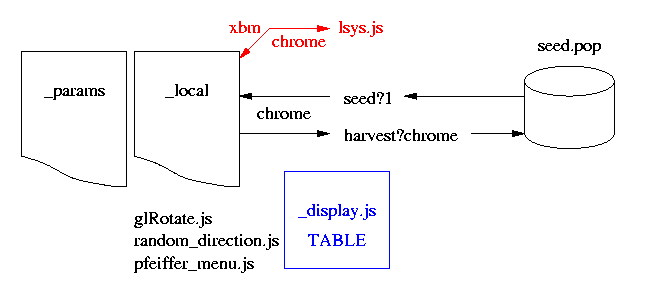
The local system is similar to the original version but has many advantages. The major differences are:
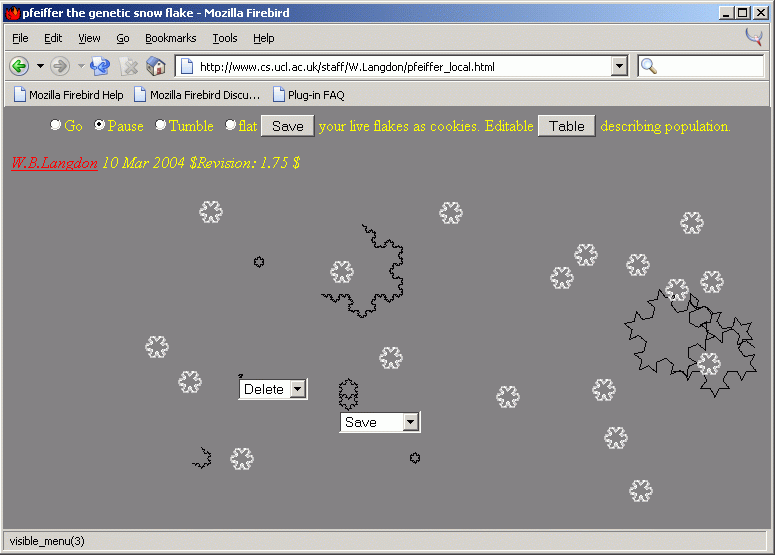 Example of main screen,
showing flying menues.
Example of main screen,
showing flying menues.
The menus appear when a mouse is dragged over a flake. This screen shot was taken, just after flake 1 (lower left) had been deleted and flake 3 (lower center) had been saved. The mouse can also be used to move flakes about the screen.
Individuals are copied, saved or deleted with the individual's menu.
The menu will appear when the simulation is paused and the mouse is
dragged across the individual.
If cookies are enabled, the cookie for that individual is updated.
Also pfeiffer_local.html
will convert the genetic material to a string of hexadecimal digits
(hexMe) and try and pass it across the network to
harvest.
An individual snow flake can be dragged and dropped elsewhere on the screen using the mouse. Allowing the user to line up potential mating pairs.
Currently (July 2003) mutation is disabled and all new individuals are created by crossover. Just as when the program starts, a new XBM bit map is created after every new individual chromosome is created. Some browsers allow colour PNG images. New babies are highlighted in light blue.
The fitness function rewards novelty and complexity. A flake produced by crossover cannot enter the population unless it is unique. (Cloning by the user, of course, ignores this requirement). Uniqueness is checked at three levels; the chromosome, graphics commands and pixel bitmap levels.
Each flake will only accept a potential mate it encounters (ie comes within mate radius of) if it appears to be of above average fitness.
From birth, each individual maintains a (noisy) estimate of the median fitness of "mature" individuals in the population. This is simply the fitness of the last "mature" guy it met. Ie only improvements are accepted.
Following mating, both parents are unable to mate for a period proportional to the cpu resources (in milliseconds) needed to create their child. (In general still born flakes which fail the uniqueness check will require little cpu time and so impose little penalty.) Note even with a small penalty, parents are unlikely to mate again immediately because they will have updated their estimates of average fitness to that of the other parent. Repeat mating is possible only if another mature flake with lower fitness is encountered.
Incest is also unlikely since a child must become mature. Random movements will tend to ensure it is far from its parents and their other offspring before it becomes mature.
The rest of the local and server based systems are similar to each other.
lsys.js etc are based on http://www.246.dk/ lsys.perl The chromosome is treated as a series of command parameters (defaults are shown in brackets):-
Maarten Keijzer and the much put upon reviewers. David Corney and Emma, Song Hu. Joao Oliveira, Mel Slater and Jano van Hemert. Jorge Tavares, Ernesto Costa, Francisco Pereira, Sara Silva.
Many URLs have been raided, including these:- Mozilla http://devedge.netscape.com http://www.cs.tut.fi/~jkorpela/forms/extraspace.html http://www.dansteinman.com/dynduo/en/mouseevents.html http://www.quirksmode.org/js/events_compinfo.html http://javascriptsource.com http://www.libpng.org/pub/png/ Phil Reeve for en Hex. http://codelifter.com/main/javascript/dragablelayer.html http://www.biglist.com/lists/xsl-list/archives/200006/msg01102.html http://www.w3.org/TR/REC-html40/interact/forms.html http://www.w3.org/TR/REC-html40/struct/tables.html
W.B.Langdon $Revision: 1.9 $ 12 April 2004 (27 Aug 2006)