one-way, three-level ANOVA (e.g. three groups)
Illustrate tests of "main effect" F-contrast and separate two-sample t-tests (standard, not repeated-measures, ANOVA)
Contents
Data
randn('state', 0); Na = 10; Nb = 10; Nc = 10; % balanced case % Na = 10; Nb = 12; Nc = 11; % unbalanced group sizes a = randn(Na, 1); b = randn(Nb, 1); c = randn(Nc, 1);
ANOVA main effect F-contrast
Y = [a;b;c];
X = [ones(size(a)) zeros(size(a)) zeros(size(a))
zeros(size(b)) ones(size(b)) zeros(size(b))
zeros(size(c)) zeros(size(c)) ones(size(c))];
Cmain = [1 -1 0
0 1 -1]';
[Fmain DFmain Bmain] = spm_ancova(X, eye(length(Y)), Y, Cmain)
ResMS = sum((Y-X*(X'*X\X'*Y)).^2)/DFmain(2)
% equivalent alternative F-contrast
Cmain2 = [1 -1/2 -1/2
-1/2 1 -1/2
-1/2 -1/2 1 ]';
[Fmain DFmain Bmain] = spm_ancova(X, eye(length(Y)), Y, Cmain)
Fmain =
0.1588
DFmain =
2 27
Bmain =
0.0013
0.2310
0.0253
ResMS =
1.0038
Fmain =
0.1588
DFmain =
2 27
Bmain =
0.0013
0.2310
0.0253
Two-sample t-contrasts in ANOVA model
Y = [a;b;c];
X = [ones(size(a)) zeros(size(a)) zeros(size(a))
zeros(size(b)) ones(size(b)) zeros(size(b))
zeros(size(c)) zeros(size(c)) ones(size(c))];
Cab = [1;-1;0];
[Tab DFab Bab] = spm_ancova(X, eye(length(Y)), Y, Cab)
Cac = [1;0;-1];
[Tac DFac Bac] = spm_ancova(X, eye(length(Y)), Y, Cac)
Tab =
-0.5126
DFab =
1.0000 27.0000
Bab =
0.0013
0.2310
0.0253
Tac =
-0.0536
DFac =
1.0000 27.0000
Bac =
0.0013
0.2310
0.0253
Separately modelled two-sample t-tests
Yab = [a;b];
Xab = [ones(size(a)) zeros(size(a))
zeros(size(b)) ones(size(b))];
Cab = [1;-1];
[Tab DFab Bab] = spm_ancova(Xab, eye(length(Yab)), Yab, Cab)
Yac = [a;c];
Xac = [ones(size(a)) zeros(size(a))
zeros(size(c)) ones(size(c))];
Cac = [1;-1];
[Tac DFac Bac] = spm_ancova(Xac, eye(length(Yac)), Yac, Cac)
Tab =
-0.5750
DFab =
1.0000 18.0000
Bab =
0.0013
0.2310
Tac =
-0.0508
DFac =
1.0000 18.0000
Bac =
0.0013
0.0253
See also anova1 and multcompare in MATLAB
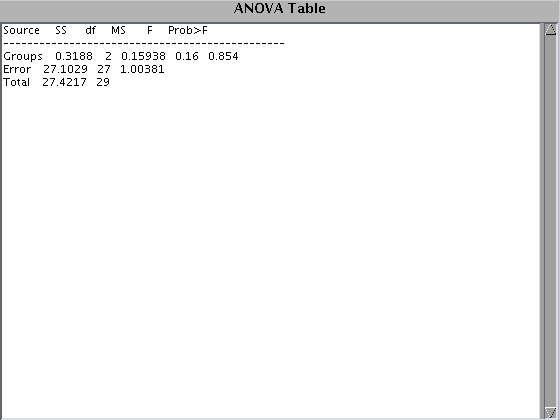
[p tab st] = anova1(Y, [ones(Na,1);2*ones(Nb,1);3*ones(Nb,1)]); st mc = multcompare(st); % Note, multcompare uses only the information in the st structure, it % doesn't perform separate two-sample t-tests. Compare e.g. Bmain = st.means' DFmain = st.df ResMS = st.s^2 % this leads me to believe that the ANOVA approach is correct. Though note % that the above didn't touch on the multiple comparison adjustment issue.
st =
gnames: {3x1 cell}
n: [10 10 10]
source: 'anova1'
means: [0.0013 0.2310 0.0253]
df: 27
s: 1.0019
Bmain =
0.0013
0.2310
0.0253
DFmain =
27
ResMS =
1.0038