Results from Genetic Programming for Combining Neural Networks for Drug Discovery
Figure 3
shows the evolved combined classifier produced by genetic
programming is better than all of the 60 trained neural networks.
It is better than
ROCs of any of the 60 neural networks.
In fact
Figure 4 shows its ROC is better than the MRROC of them all together.
(The
movie
shows the evolution of the ROCs in the GP population
in a similar run).
Graph (purple) shows
size fair crossover and the 4 mutation operators have
succeeded in controlling bloat.
The evolved classifier is shown at the end.
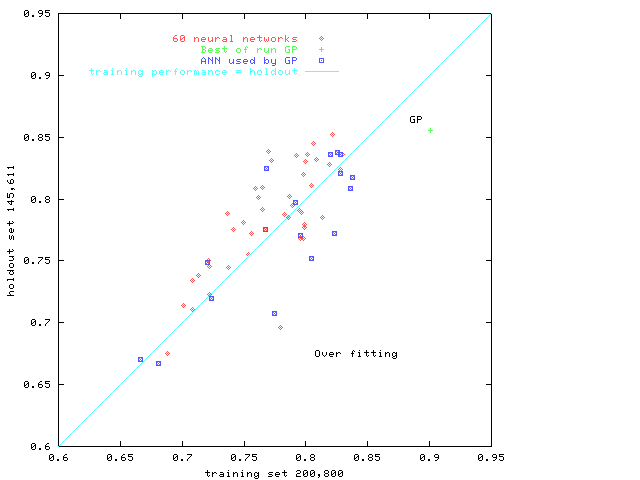
Fig. 3.
Performance of 60 given neural networks
and genetic programming.
The area under the
Receiver Operating Characteristics (ROC)
is plotted on the training data (horizontal)
versus
holdout data (vertical).
Points below the diagonal indicate a degree of over training.
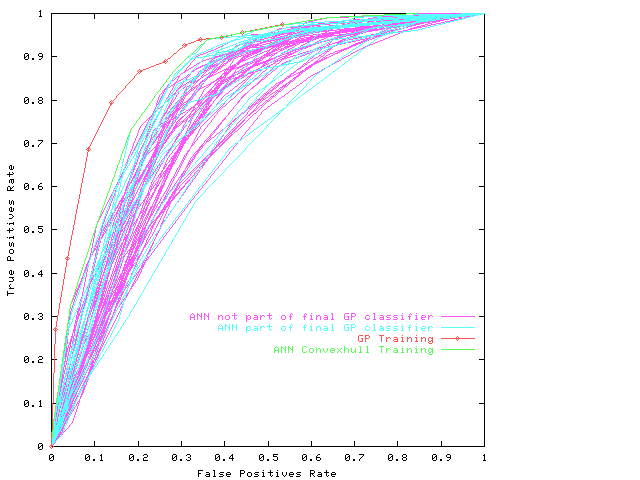
Receiver Operating Characteristics
on the training data
of evolved composite classifier.
For comparison the ROCs of 60 given neural networks
are also plotted.
The 17 that are included in the final
are in different colour.
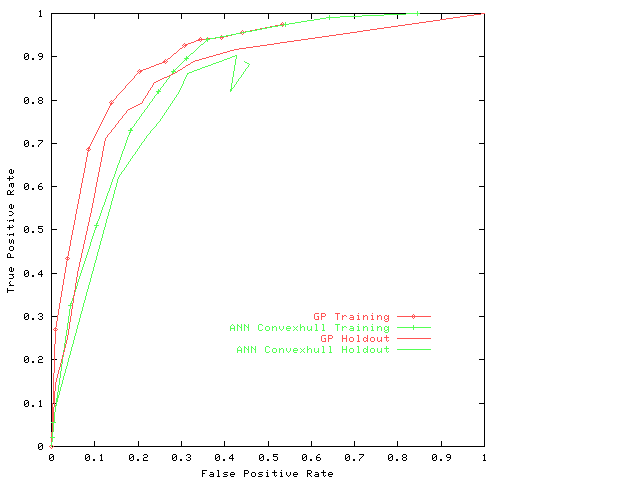
Fig. 4.
Receiver Operating Characteristics
of evolved composite classifier.
For comparison the convex hull of the 60 given neural networks,
on the training and holdout data, is given.
Note the convex hull classifier is no longer convex when
used to classify the holdout data.
Movie
showing the evolution of individual classifiers within the GP
population plotted in the ROC square.
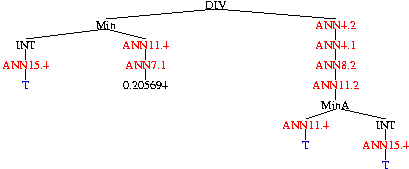
Evolved P450 Classifier
 |
| Tree 0 |
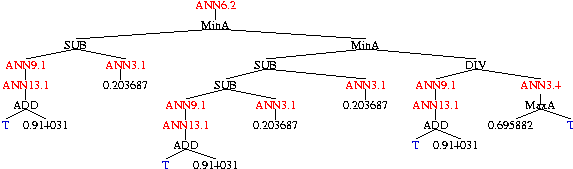 |
| Tree 1 |
 |
| Tree 2 |
 |
| Tree 3 |
 |
| Tree 4 |
Five combinations are evolved simultaneously.
Each consists of the neural networks trained by Clementine
(in red),
arithmetic functions,
constants
and the tuning (or sensitivity) parameter T (in blue).
The complete classifier is obtained by summing the outputs of the five
trees.
If the sum is negative, this predicts the chemical will be inactive.
The Function set
- floating point +, -, times, and divide
Note however divide by zero
always yields 1.0.
This protects it and prevents the GP system failing with a
divide-by-zero fault.
- Max takes two arguments and returns the value of the
largest.
- Min returns the value of the smaller
- MaxA also takes two arguments but returns the signed value of the
largest in absolute terms.
E.g. MaxA (-2,1) returns -2.0
- MinA returns the signed value of the
smallest in absolute terms.
- E.g. INT(3.23) returns 3.
- FRAC returns the fractional part of its input.
E.g. FRAC(3.23) returns 0.23.
- ANNxx.y
is the output of the neural network trained on featureset group xx
and dataset y.
Its argument is its tuning parameter.
A value of 0.5 indicates no baise.
Using the evolved classifier
When presented with a chemical to be classified.
Each of the neural networks (in the evolved composite classifier)
makes its own estimate of whether the chemical is active or not.
To do this it needs about 50 inputs.
(which ones depend upon which of 15 groups the neural network belongs
to).
The inputs are properties of the chemical.
There are a total of 699 chemical features (inputs).
These have been split into 15 groups of about 50.
The evolved classifier uses 17 neural networks
but some come from the same group.
In fact only 12 groups are used.
So approximately 560 features are need for the chemical.
The outputs of the neural networks are
then treated as implicit inputs by the five evolved trees.
The value returned internally within the GP calculation
depend both on the threshold argument to the ANN function and
(implicitly) on the output of the neural network.
Additionally the tuning parameter T must be set
to a value in the range 0 to 1.
Note each GP tree makes a nonlinear combination of the results given
by several neural networks.
Finally the output of all five trees are summed.
If the value is non-negative,
this means the chemical is predicted to be active against the specific
P450 enzyme.
back
W.B.Langdon@cs.ucl.ac.uk
30 August 2001







