GECCO 2004 LATE BREAKING PAPER
Repeated Sequences in Linear GP Genomes
W. B. Langdon
and
W. Banzhaf
Computer Science,
Memorial University of Newfoundland, Canada

W.Langdon@cs.uc1.ac.uk, banzhaf@cs.mun.ca
50% of your DNA is Repeated Sequences
How much of your programs is Repeated Sequences?
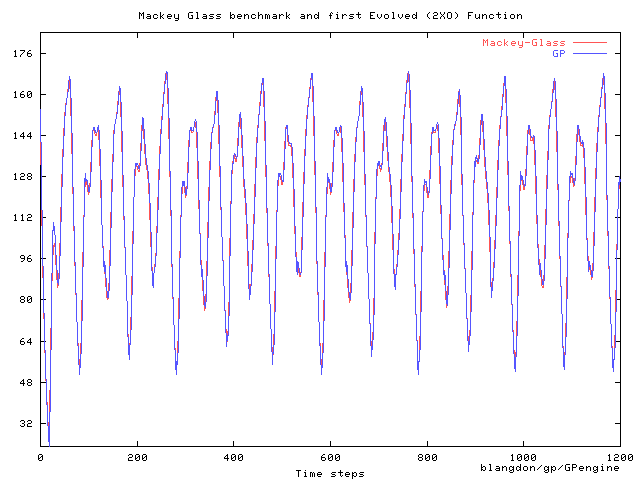
Discrete Mackey-Glass chaotic time series
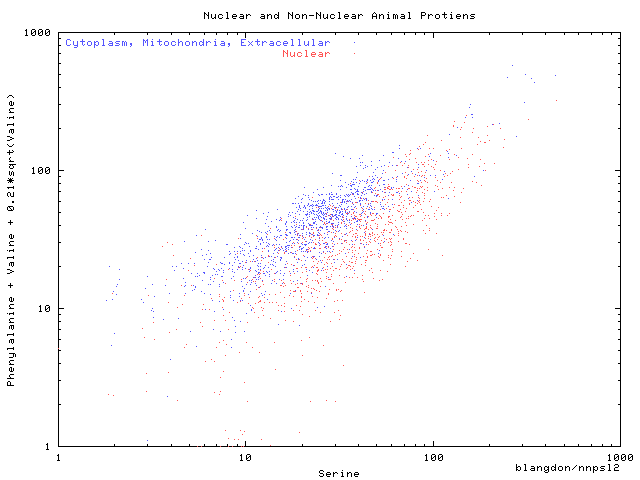
The three (of twenty) amino acids plotted where selected
by Discipulus as being the best discriminators.
The non-linear function of number of Valines
was also suggested by an evolved model.
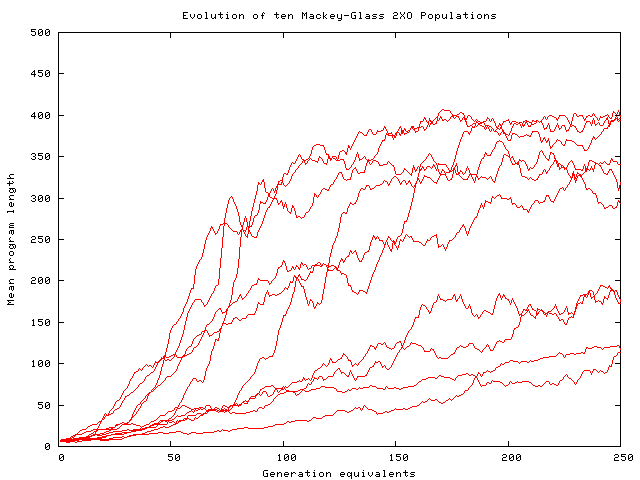
Evolution of mean program size
in ten Mackey-Glass prediction runs
using two point crossover.
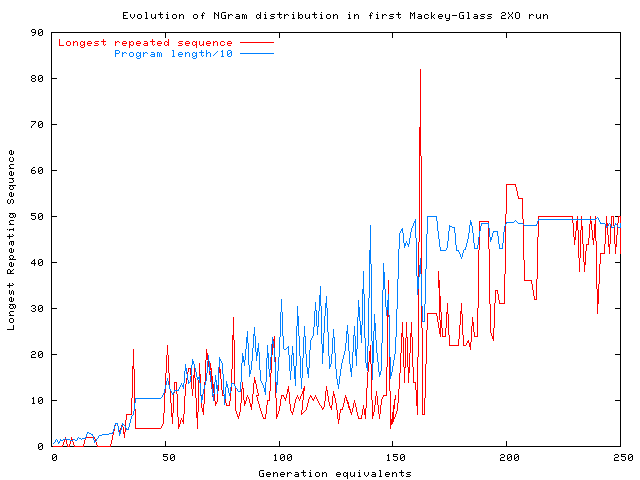
Evolution of length of longest repeated sequence of instructions in
the best Mackey-Glass prediction program produced by
the
first run with
two-point crossover (2XO) and fitness selection.
The length of the programs is also shown.
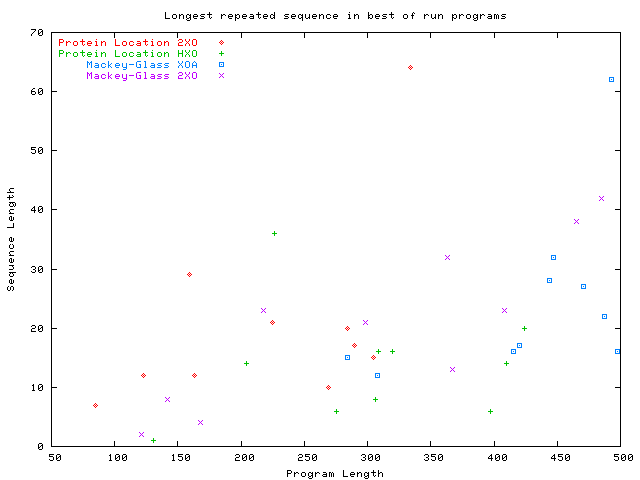
Length of longest repeated sequence of instructions in
the best prediction programs.
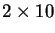 Discipulus protien locations runs
and
Discipulus protien locations runs
and
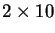 Mackey-Glass prediction runs.
All bloating runs evolve repeated sequences.
Mackey-Glass prediction runs.
All bloating runs evolve repeated sequences.
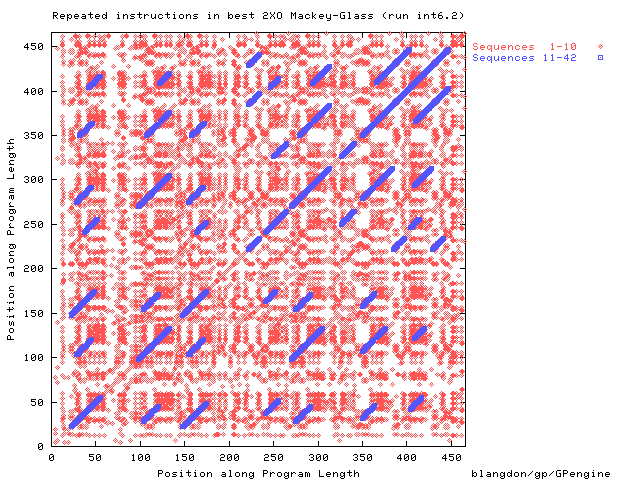
Location of repeated instructions in
the best Mackey-Glass prediction program.
Instructions that are part of
repeated sequences longer than 10 are plotted in blue.
Notice almost every instruction is repeated,
so the diagonal is almost solid.
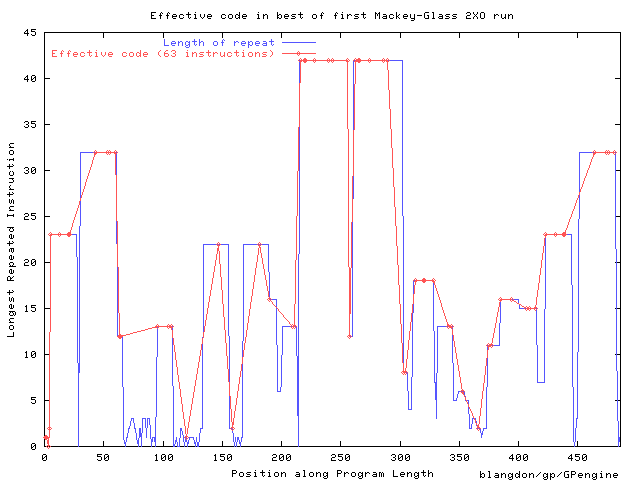
Distribution of repeated sequences along length of best
Mackey-Glass
predictor at
end of first 2XO run.
The solid line highlights the location of its 63 effective
instructions.
An animation can be found via
http://www.cs.ucl.ac.uk/staff/W.Langdon/gecco2004lb/
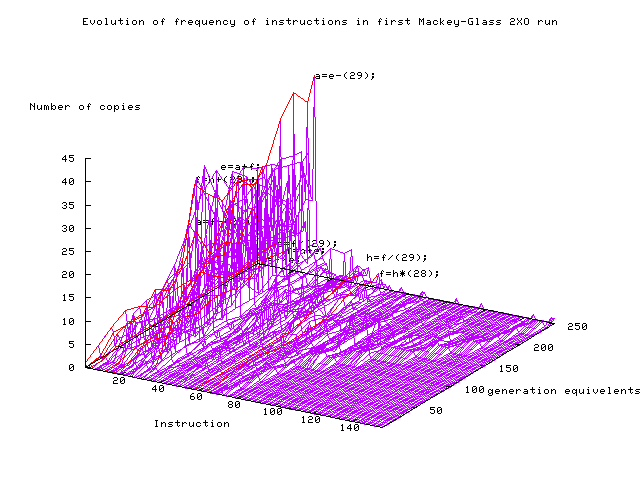
Evolution of instructions in
first 2XO Mackey-Glass run.
The red lines and text highlight the effective
instructions
(e.g. a=e-(29);)
in the best of run program.
Bill LANGDON
2004-07-07