|
Corresponding author: Prof. Natasa Przulj, e-mail: natasa [AT] cs.ucl.ac.uk
- About:
- Executables:
- Networks:
- Alignments:
- Predictions:
How to run C-GRAAL:
- Download and unpack the C-GRAAL using the link above.
- C-GRAAL accepts networks in LEDA .gw format.
- Run "./CGRAAL_unix64" without any parameters and it will display usage directions
- Usage directions:"./CGRAAL_unix64 net1.gw net2.gw similarity.file output1.file output2.file", where:
- "net1.gw" - first network (gw format)
- "net2.gw" - second network (gw format)
- "similarity.file": similarity between nodes in net1 and net2 in the following format: nodeXNet1 nodeYNet2 similarity
- "output1.file"
- alignment output file in following format: node_Net1 (aligned to)
node_Net2, where nodes are represented as node numbers
- "output2.file"
- alignment output file in following format: node_Net1 (aligned to)
node_Net2, where nodes are represented as node names
- Note: Number of nodes in the first network must not be greater than number of nodes in the second network!
How to convert network from edge list format to .gw file accepted by C-GRAAL:
If your network is in edge list format:
node1 node2
node3 node4
...
You can use "list2leda" script (part of "CGRAAL_unix64.zip") to convert it into the format accepted by C-GRAAL.
Use it like this: "./list2leda edgelist.txt >> graph.gw"
Note that self-loops, directionality and double edges will be ignored.
Figures:
 |  |  |
|---|
The largest common connected component of the best yeast-human alignment | The largest common connected component of the best C. jejuni-E. coli alignment | The largest common connected component of the best Mesorhizobium loti-Synechocystis sp PCC6803 alignment |
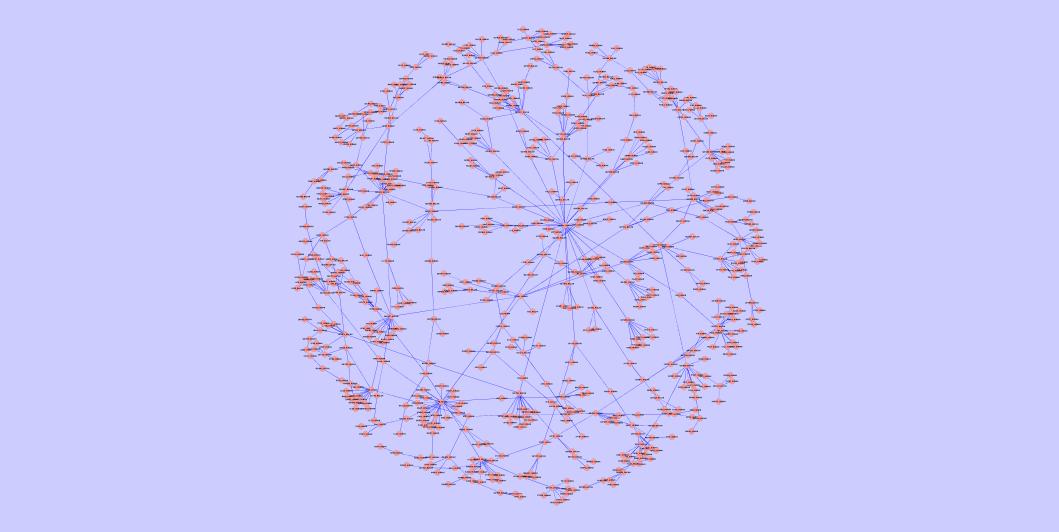 | 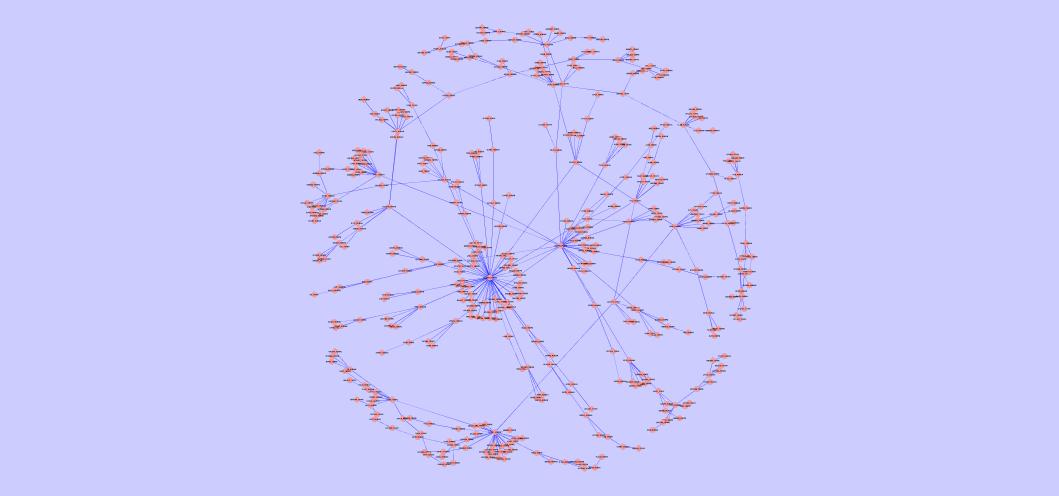 | 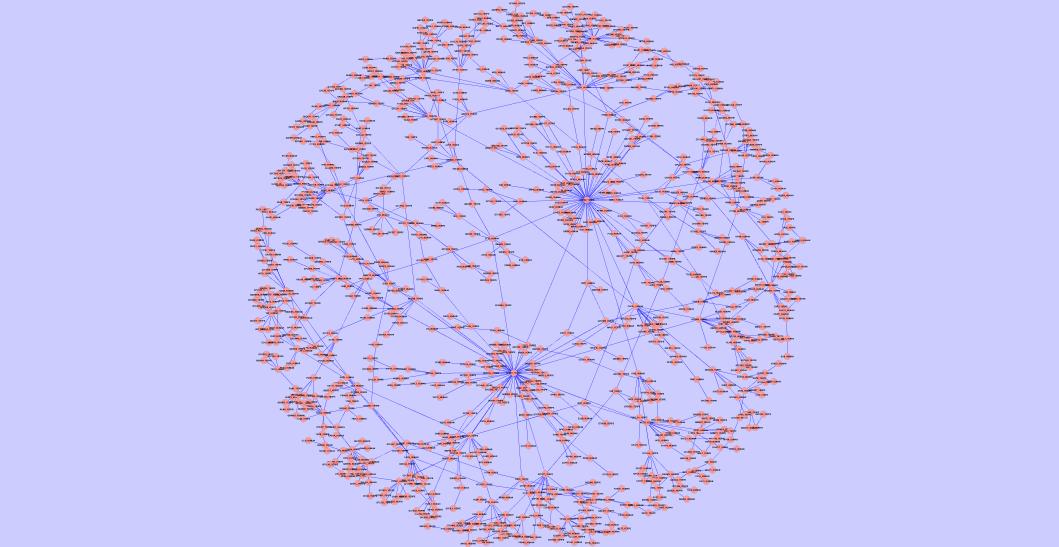 |
|---|
The largest common connected component of the best F.tularensis - H.sapiens alignment | The largest common connected component of the best B.anthracis - H.sapiens alignment | The largest common connected component of the best Y.pestis - H.sapiens alignment |
Other network alignment algorithms:
Notes:- All
p-values presented in this paper were calculated using Matlab 2007.
Newer Matlab versions use a different output precision format
and
consequently, it is possible that the p-values obtained by the
newer versions will differ from those obtained by the version 2007
(e.g., old version p-value: 1.7E-9, newer version p-value: 0.0).
|



